API v2, New BDE Methods, MCP, And More
new API philosophy; streamlined interfaces for workflows; using NNPs and g-xTB to predict bond strength; an MCP server; .sdf files; benchmarking protein–ligand interactions; Diels–Alder visualizations
We’re excited to share a set of major improvements to our API today, along with a host of interesting features, improvements, and benchmarks that we’ve been working on.
Rowan’s v2 Python API
Since launching our first programmatic interface, tens of thousands of workflows have been run through Rowan’s API, automating complex scientific tasks and accelerating research. The feedback and insights from that usage have been invaluable, and we’ve worked hard to incorporate all suggestions into our next major release.
Today, we're excited to launch Rowan API v2. This is a significant update to our Python library, designed from the ground up to make Rowan’s API more intuitive, consistent, and powerful.
What’s New in v2?
Functions and Objects Working Together
The most fundamental change in v2 is a shift in our design philosophy. We've moved from an approach that relied exclusively on class methods to a hybrid model that combines the strengths of object-oriented and functional programming.
For simple, direct actions, you can now use standalone functions. When you need to inspect or work with results, you get back powerful objects with their own useful methods. This makes common tasks cleaner while preserving deeper functionality for when you need it.
See the difference for yourself. Submitting a workflow and retrieving the result now looks like this:
# v1 approach
import rowan
rowan.api_key = "rowan-sk-..."
submission = rowan.Workflow.submit(...)
result = rowan.Workflow.retrieve(submission["uuid"])
# then parse the result dictionary
print(result["object_status"])
# v2 approach
import rowan
rowan.api_key = "rowan-sk-..."
result = rowan.submit_workflow(...)
# The result is an object with methods
result.fetch_latest(in_place=True)
print(result.status)This new pattern reduces the amount of boilerplate needed for routine scripting, giving you the simplicity of functions for inputs and the power of objects for outputs.
First-Class Functions for Workflows
API v2 introduces first-class functions for our core scientific workflows. This gives you direct, named access to Rowan’s powerful computational engines. Whether you’re predicting pKa values, running conformational searches, or modeling protein–ligand binding, v2 provides dedicated functions that reduce the need for manual parameter configuration.
For example, running a pKa calculation is now much more direct:
# v1 approach
import rowan
import stjames
rowan.api_key = "rowan-sk-..."
result = rowan.Workflow.submit(
workflow_type="pka",
workflow_data={
"pka_range": (2,12),
"deprotonate_elements": [7, 8, 16],
"protonate_elements": [7],
"mode": "careful",
},
initial_molecule=stjames.Molecule.from_smiles("O")
)
# v2 approach
import rowan
import stjames
rowan.api_key = "rowan-sk-..."
result = rowan.submit_pka_workflow(
initial_molecule=stjames.Molecule.from_smiles("n1ccccc1"),
mode="careful",
name="Pyridine pKa",
)These new functions come with sane defaults that mirror our web application, but remain tunable for advanced use cases.
Structured Returns, Not Raw JSON
We're moving on from raw JSON returns. In v2, the API returns rich, Pydantic-backed Python objects for everything from folders and proteins to workflows and your user account.
This is a huge improvement to the developer experience, giving you:
IDE Autocompletion: No more guesswork or checking documentation for dictionary keys.
Static Type Checking: Catch errors in your code before you even run it.
Cleaner, Self-Documenting Code:
result.statusis always better thanresult['status'].
Cofolding
Protein cofolding with the popular Chai and Boltz models was previously only available through the web interface. We're excited to bring this powerful capability to the API in v2. Running these workflows is now as simple as a single function call:
result = rowan.submit_protein_cofolding_workflow(
initial_protein_sequences=[ "ASKGTSHEAGIVCRITKPALLVLNHETAKVIQTAFQRASYPDITGEKAMMLLGQVKYGLHNIQISHLSIASSQVELVEAKSIDVSIQDVSVVFKGTLKYGYTTAWWLGIDQSIDFEIDSAIDLQINTQLTADSGRVRTDAPDCYLSFHKLLLHLQGEREPGWIKQLFTNFISFTLKLVLKGQICKEINVISNIMADFVQTRAASILSDGDIGVDISLTGDPVITASYLESHHKGHFIYKDVSEDLPLPTFSPTLLGDSRMLYFWFSERVFHSLAKVAFQDGRLMLSLMGDEFKAVLETWGFNTNQEIFQEVVGGFPSQAQVTVHCLKMPKISCQNKGVVVDSSVMVKFLFPRPDQQHSVAYTFEEDIVTTVQASYSKKKLFLSLLDFQITPKTVSNLTESSSESIQSFLQSMITAVGIPEVMSRLEVVFTALMNSKGVSLFDIINPEIITRDGFLLLQMDFGFPEHLLVDFLQSLS"
],
initial_smiles_list=[
"CCOC(=O)N1c2ccc(C(F)(F)F)cc2[C@@H](N(Cc2cc(C(F)(F)F)cc(C(F)(F)F)c2)C(=O)OC)C[C@H]1CC"
],
ligand_binding_affinity_index=0,
name="Torcetrapib Cofolding",
)Backwards Compatibility
We know that migrating can be a painful process, especially for those with complex workflows built on v1. The original API will continue to be fully supported. You can pin your rowan-python version to continue using the v1 interface without interruption.
Try It Today
Rowan API v2 is available now. Here’s how to get started:
Install the library:
pip install --upgrade rowan-python
Explore the Docs: Our API documentation has been fully updated for v2, and you can find detailed examples, like running a high-throughput screen for a library of compounds or predicting the solubility of a PROTAC, on our GitHub.
Sign Up: If you're new to Rowan, you can create a free account at labs.rowansci.com to get an API key. All users get first-class API access, no payment required.
As always, we’d love your feedback. If something’s confusing, broken, or just feels awkward, please don’t hesitate to reach out or open an issue on GitHub.
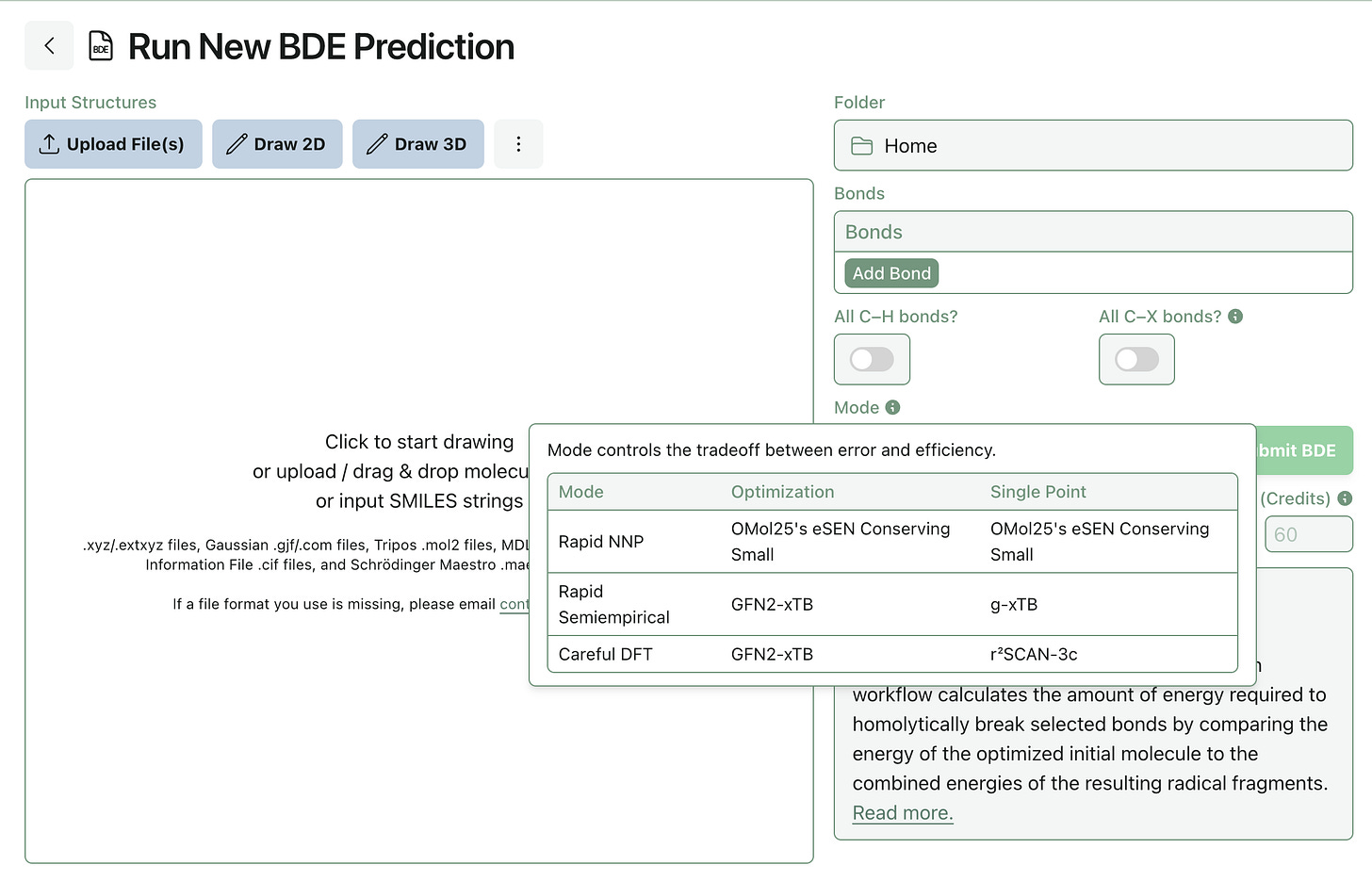
Improvements to BDE Prediction
Jonathon recently tested the ability of low-cost computational methods to reproduce a benchmark set of experimentally measured BDE values. He found that eSEN-OMol25-sm-conserving and g-xTB were Pareto-optimal, giving similar accuracy to DFT calculations at a fraction of the computational cost. (You can read the full paper here.)
Rowan subscribers can now run these low-cost BDE calculations through our bond-dissociation-energy workflow:
An MCP Server For Rowan
Model Context Protocol (MCP), developed recently by Anthropic, allows LLMs to use external tools to answer tricky questions, just like Deep Research can search the web to help find data. Kat Yenko recently created an open-source MCP server for Rowan, allowing Claude and other models to use Rowan’s API to simulate molecular properties and supplementing their native chemical intuition.
Here’s what this looks like in practice:
If you’re interested in the intersection of agentic AI and scientific computing, check this out! (You can find Kat’s original posts on X and LinkedIn.)
.sdf Download
We recently added the ability to download molecular structures as .sdf files, making it easier to go back and forth between Rowan and other drug-design software. Please let us know if we’re missing support for other useful file formats!
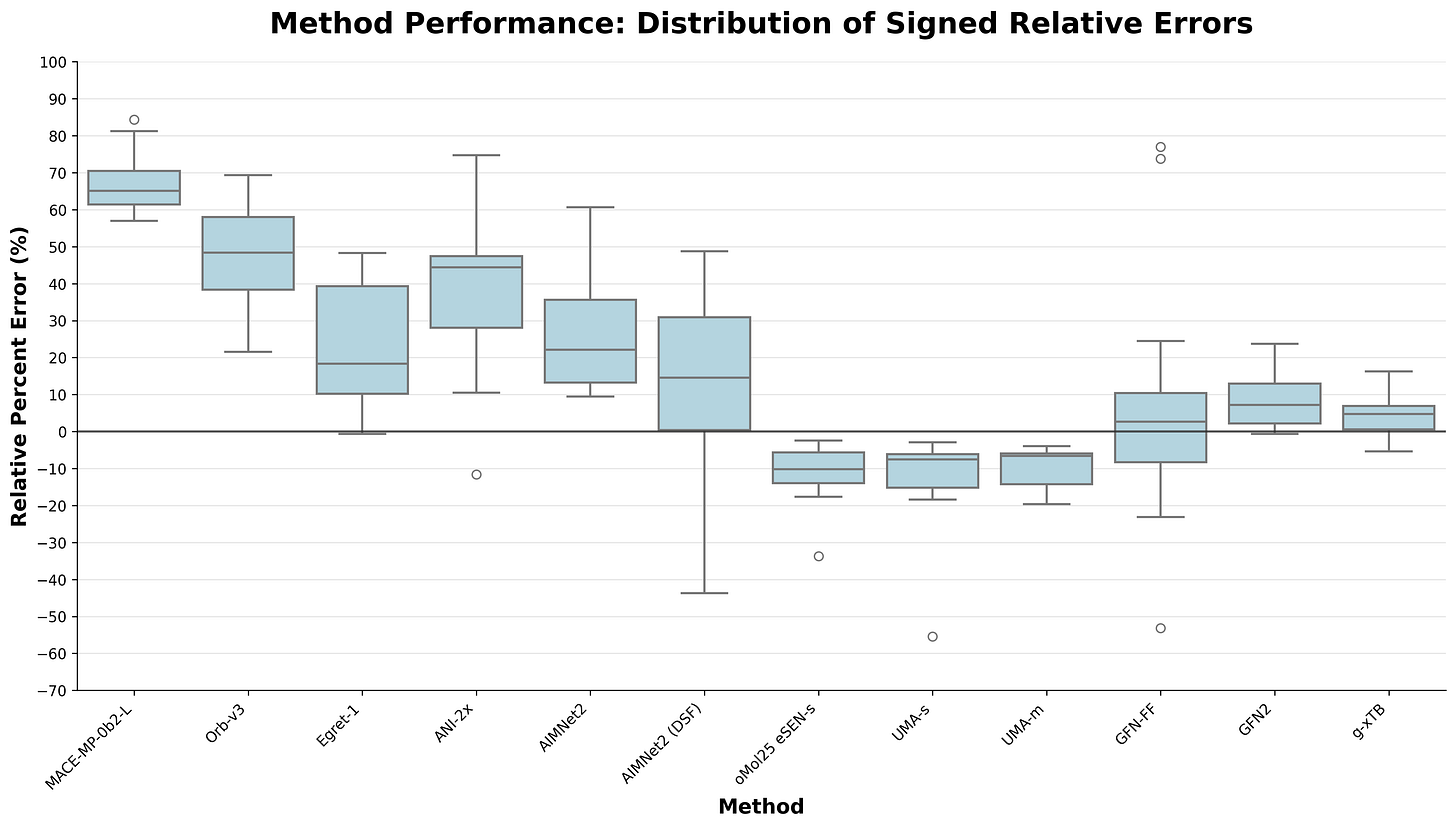
Benchmarking Protein–Ligand Interactions
Our summer intern Ishaan recently benchmarked a variety of neural network potentials on a set of 15 high-level protein–ligand interaction energies. He found that, while some NNPs were worse than others, g-xTB and GFN2-xTB still perform best:
(Corin presented these results on his poster at the Computer-Assisted Drug Design Gordon Research Conference this past week—thanks for those of you who stopped by to discuss!)
Interactive Diels–Alder Visualizations
Finally, our summer intern Isaiah created some awesome 3D visualizations of a Diels–Alder potential-energy surface using Manim, the software used in 3Blue1Brown's explainer videos:
We’re hoping to make more visualizations and incorporate these into lessons or labs for the classroom. If you’re interested in using these in your class, please reach out!